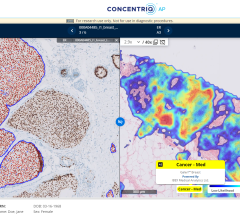
June 28, 2016 — Pathologists have been largely diagnosing disease the same way for the past 100 years, by manually reviewing images under a microscope. But new work suggests that computers can help doctors improve accuracy and significantly change the way cancer and other diseases are diagnosed.
A research team from Beth Israel Deaconess Medical Center (BIDMC) and Harvard Medical School (HMS) recently developed artificial intelligence (AI) methods aimed at training computers to interpret pathology images, with the long-term goal of building AI-powered systems to make pathologic diagnoses more accurate.
“Our AI method is based on deep learning, a machine-learning algorithm used for a range of applications including speech recognition and image recognition,” explained pathologist Andrew Beck, M.D., Ph.D., director of bioinformatics at the Cancer Research Institute at BIDMC and an associate professor at Harvard Medical School. “This approach teaches machines to interpret the complex patterns and structure observed in real-life data by building multi-layer artificial neural networks, in a process which is thought to show similarities with the learning process that occurs in layers of neurons in the brain’s neocortex, the region where thinking occurs.”
Watch the VIDEO “Examples of Artificial Intelligence in Medical Imaging Diagnostics.” This shows an example of how AI can assess mammography images.
Watch the VIDEO “Development of Artificial Intelligence to Aid Radiology,” an interview with Mark Michalski, M.D., director of the Center for Clinical Data Science at Massachusetts General Hospital, explaining the basis of artificial intelligence in radiology.
The Beck lab’s approach was recently put to the test in a competition held at the annual meeting of the International Symposium of Biomedical Imaging (ISBI), which involved examining images of lymph nodes to decide whether or not they contained breast cancer. The research team of Beck and his lab’s post-doctoral fellows Dayong Wang, Ph.D., and Humayun Irshad, Ph.D., and student Rishab Gargya, together with Aditya Khosla of the Massachusetts Institute of Technology (MIT) Computer Science and Artificial Intelligence Laboratory, placed first in two separate categories, competing against private companies and academic research institutions from around the world. The research team posted a technical report describing their approach to the arXiv.org repository, an open access archive of e-prints in physics, mathematics, computer science, quantitative biology, quantitative finance and statistics.
“Identifying the presence or absence of metastatic cancer in a patient’s lymph nodes is a routine and critically important task for pathologists,” Beck explained. “Peering into the microscope to sift through millions of normal cells to identify just a few malignant cells can prove extremely laborious using conventional methods. We thought this was a task that the computer could be quite good at – and that proved to be the case.”
In an objective evaluation in which researchers were given slides of lymph node cells and asked to determine whether or not they contained cancer, the team’s automated diagnostic method proved accurate approximately 92 percent of the time, explained Khosla, adding, “This nearly matched the success rate of a human pathologist, whose results were 96 percent accurate.”
“But the truly exciting thing was when we combined the pathologist’s analysis with our automated computational diagnostic method, the result improved to 99.5 percent accuracy,” said Beck. “Combining these two methods yielded a major reduction in errors.”
The team trained the computer to distinguish between cancerous tumor regions and normal regions based on a deep multilayer convolutional network.
“In our approach, we started with hundreds of training slides for which a pathologist has labeled regions of cancer and regions of normal cells,” said Wang. “We then extracted millions of these small training examples and used deep learning to build a computational model to classify them.” The team then identified the specific training examples for which the computer is prone to making mistakes and re-trained the computer using greater numbers of the more difficult training examples. In this way, the computer’s performance continued to improve.
“There have been many reasons to think that digitizing images and using machine learning could help pathologists be faster, more accurate and make more accurate diagnoses for patients,” Beck added. “This has been a big mission in the field of pathology for more than 30 years. But it’s been only recently that improved scanning, storage, processing and algorithms have made it possible to pursue this mission effectively. Our results in the ISBI competition show that what the computer is doing is genuinely intelligent and that the combination of human and computer interpretations will result in more precise and more clinically valuable diagnoses to guide treatment decisions."
Jeroen van der Laak, Ph.D., who leads a digital pathology research group at Radboud University Medical Center in the Netherlands and was an organizer for the competition, said, “When we started this challenge, we expected some interesting results. The fact that computers had almost comparable performance to humans is way beyond what I had anticipated. It is a clear indication that artificial intelligence is going to shape the way we deal with histopathological images in the years to come.”
Beck and Khosla recently formed a start-up company (PathAI) with the mission of developing and applying AI technology for pathology.
For more information: www.bidmc.org


 April 17, 2025
April 17, 2025